Simple and multiple regression#
Show code cell content
import numpy as np
import pandas as pd
# Safe setting for Pandas. Needs Pandas version >= 1.5.
pd.set_option('mode.copy_on_write', True)
import matplotlib.pyplot as plt
plt.style.use('fivethirtyeight')
np.set_printoptions(suppress=True)
Show code cell content
# For calculating correlation
def standard_units(any_numbers):
""" Convert any array of numbers to standard units.
"""
return (any_numbers - np.mean(any_numbers))/np.std(any_numbers)
def correlation(t, x, y):
""" Correlation of columns `x` and `y` from data frame `t`
"""
return np.mean(standard_units(t[x]) * standard_units(t[y]))
Show code cell content
def scatter_errors(x_values, y_values, c, s):
""" Plot a line through data with errors
Parameters
----------
x_values : array
Values we are predicting from, for the x-axis of the plot.
y_values : array
Values we are predicting, for the y-axis of the plot.
c : number
Intercept for predicting line.
s : number
Slope for predicting line.
Returns
-------
rmse : number
The square root of the mean squared error, for the given `x_values`,
`y_values` and line.
"""
# Predict the y values from the line.
predicted = c + s * x_values
# Errors are the real values minus the predicted values.
errors = y_values - predicted
# Plot real values in blue, predicted values in red.
actual_points = plt.plot(x_values, y_values, 'o', color='blue')
predicted_points = plt.plot(x_values, predicted, 'o', color='red')
# Draw a line between predicted and actual
for i in np.arange(len(x_values)):
x = x_values[i]
y_0 = predicted[i]
y_1 = y_values[i]
error_line = plt.plot([x, x], [y_0, y_1], ':', color='black', linewidth=1)
plt.legend(actual_points + predicted_points + error_line,
['Actual', 'Predicted', 'Error'])
return np.sqrt(np.mean(errors ** 2))
Simple and multiple regression#
We looked at simple regression in the finding lines page, and those following.
Simple regression uses a single set of predictor values, and a straight line, to predict another set of values.
For example, in the finding lines page above, we predicted the “quality” scores (on the y-axis) from the “easiness” scores (on the x-axis).
This page is about multiple regression. Multiple regression takes simple regression a step further. Now we use more than one set of values to predict another set of values.
On the way, we will start using a standard statistics library in Python, called StatsModels.
Simple regression#
Let us return to simple regression — using one set of values (on the x axis) to predict another set of values (on the y axis).
Here is our familiar chronic kidney disease dataset.
ckd = pd.read_csv('data/ckd_clean.csv')
ckd
| Age | Blood Pressure | Specific Gravity | Albumin | Sugar | Red Blood Cells | Pus Cell | Pus Cell clumps | Bacteria | Blood Glucose Random | ... | Packed Cell Volume | White Blood Cell Count | Red Blood Cell Count | Hypertension | Diabetes Mellitus | Coronary Artery Disease | Appetite | Pedal Edema | Anemia | Class | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 48.0 | 70.0 | 1.005 | 4.0 | 0.0 | normal | abnormal | present | notpresent | 117.0 | ... | 32.0 | 6700.0 | 3.9 | yes | no | no | poor | yes | yes | 1 |
| 1 | 53.0 | 90.0 | 1.020 | 2.0 | 0.0 | abnormal | abnormal | present | notpresent | 70.0 | ... | 29.0 | 12100.0 | 3.7 | yes | yes | no | poor | no | yes | 1 |
| 2 | 63.0 | 70.0 | 1.010 | 3.0 | 0.0 | abnormal | abnormal | present | notpresent | 380.0 | ... | 32.0 | 4500.0 | 3.8 | yes | yes | no | poor | yes | no | 1 |
| 3 | 68.0 | 80.0 | 1.010 | 3.0 | 2.0 | normal | abnormal | present | present | 157.0 | ... | 16.0 | 11000.0 | 2.6 | yes | yes | yes | poor | yes | no | 1 |
| 4 | 61.0 | 80.0 | 1.015 | 2.0 | 0.0 | abnormal | abnormal | notpresent | notpresent | 173.0 | ... | 24.0 | 9200.0 | 3.2 | yes | yes | yes | poor | yes | yes | 1 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 153 | 55.0 | 80.0 | 1.020 | 0.0 | 0.0 | normal | normal | notpresent | notpresent | 140.0 | ... | 47.0 | 6700.0 | 4.9 | no | no | no | good | no | no | 0 |
| 154 | 42.0 | 70.0 | 1.025 | 0.0 | 0.0 | normal | normal | notpresent | notpresent | 75.0 | ... | 54.0 | 7800.0 | 6.2 | no | no | no | good | no | no | 0 |
| 155 | 12.0 | 80.0 | 1.020 | 0.0 | 0.0 | normal | normal | notpresent | notpresent | 100.0 | ... | 49.0 | 6600.0 | 5.4 | no | no | no | good | no | no | 0 |
| 156 | 17.0 | 60.0 | 1.025 | 0.0 | 0.0 | normal | normal | notpresent | notpresent | 114.0 | ... | 51.0 | 7200.0 | 5.9 | no | no | no | good | no | no | 0 |
| 157 | 58.0 | 80.0 | 1.025 | 0.0 | 0.0 | normal | normal | notpresent | notpresent | 131.0 | ... | 53.0 | 6800.0 | 6.1 | no | no | no | good | no | no | 0 |
158 rows × 25 columns
In our case, we restrict ourselves to the chronic kidney disease patients.
These patients have a 1 in the Class column.
We’re also going to restrict ourselves to looking at the following measures:
Serum Creatinine: a measure of how well the kidney is clearing substances from the blood. When creatinine is high, it means the kidney is not clearing well. This is the general measure of kidney disease that we are interested to predict.Blood Urea: another measure of the ability of the kidney to clear substances from the blood. Urea is high in the blood when the kidneys are not clearing efficiently.Hemoglobin: healthy kidneys release a hormone erythropoietin that stimulates production of red blood cells, and red blood cells contain the hemoglobin molecule. When the kidneys are damaged, they produce less erythropoietin, so the body produces fewer red blood cells, and there is a lower concentration of hemoglobin in the blood.White Blood Cell Count: white cells are the immune cells in the blood. White cells increase in number when there is some inflammatory process in the body. There is some dispute about whether the white blood cell count is a useful measure in chronic kidney disease.
# Data frame restricted to kidney patients and columns of interest.
ckdp = ckd.loc[
ckd['Class'] == 1, # Kidney disease patients.
['Serum Creatinine', # Columns of interest.
'Blood Urea',
'Hemoglobin',
'White Blood Cell Count']]
# Rename the columns with shortened names.
ckdp.columns = ['Creatinine', 'Urea', 'Hemoglobin', 'WBC']
ckdp.head()
| Creatinine | Urea | Hemoglobin | WBC | |
|---|---|---|---|---|
| 0 | 3.8 | 56.0 | 11.2 | 6700.0 |
| 1 | 7.2 | 107.0 | 9.5 | 12100.0 |
| 2 | 2.7 | 60.0 | 10.8 | 4500.0 |
| 3 | 4.1 | 90.0 | 5.6 | 11000.0 |
| 4 | 3.9 | 148.0 | 7.7 | 9200.0 |
First let us look at the relationship of the urea levels and the creatinine:
ckdp.plot.scatter('Urea', 'Creatinine')
<Axes: xlabel='Urea', ylabel='Creatinine'>

There is a positive correlation between these sets of values; high urea and high creatinine go together; both reflect the failure of the kidneys to clear those substances from the blood.
correlation(ckdp, 'Urea', 'Creatinine')
np.float64(0.8458834154017618)
Now recall our standard method of finding a straight line to match these two
attributes, where we choose our straight line to minimize the root mean squared
error between the straight line prediction of the Creatinine values from the
Urea values, and the actual values of Creatinine.
def rmse_any_line(c_s, x_values, y_values):
""" Root mean squared error for intercept, slope `c_s`
"""
c, s = c_s
predicted = c + x_values * s
error = y_values - predicted
return np.sqrt(np.mean(error ** 2))
We find the least-(root mean) squares straight line, using an initial guess for
the slope and intercept of [0, 0].
Again we use the Powell method to search for the minimum.
from scipy.optimize import minimize
initial_guess = [0, 0]
min_res = minimize(rmse_any_line,
initial_guess,
args=(ckdp['Urea'], ckdp['Creatinine']),
method='powell')
min_res
message: Optimization terminated successfully.
success: True
status: 0
fun: 2.2218544954119746
x: [-8.491e-02 5.524e-02]
nit: 3
direc: [[ 0.000e+00 1.000e+00]
[-4.226e+00 2.939e-02]]
nfev: 74
In particular, our intercept and slope are:
min_res.x
array([-0.0849111 , 0.05524186])
You have already seen for this special case, of the root mean square (or the
sum of squares) error, we can get the same answer directly with calculation. We
used linregress from scipy.stats to do this calculation in earlier pages.
from scipy.stats import linregress
linregress(ckdp['Urea'], ckdp['Creatinine'])
LinregressResult(slope=np.float64(0.05524183911456146), intercept=np.float64(-0.08491111825351982), rvalue=np.float64(0.845883415401762), pvalue=np.float64(9.327350567383254e-13), stderr=np.float64(0.005439919989116022), intercept_stderr=np.float64(0.6680061444061487))
Notice that the slope and the intercept are the same as those from minimize
above, within the precision of the calculation, and that the rvalue above is
the same as the correlation:
correlation(ckdp, 'Urea', 'Creatinine')
np.float64(0.8458834154017618)
StatsModels#
Now it is time to introduce a major statistics package in Python, StatsModels.
StatsModels does many statistical calculations; among them are simple and multiple regression. Statsmodels categorizes these types of simple linear models as “ordinary least squares” (OLS).
Here we load the StatModels interface that uses Pandas data frames:
# Get the Pandas interface to the StatsModels routines.
import statsmodels.formula.api as smf
Next we specify our model using a formula. Read the ~ in the formula below
as “as a function of”. So the formula specifies a linear (straight-line) model
predicting Creatinine as a function of Urea.
simple_model = smf.ols(formula="Creatinine ~ Urea", data=ckdp)
Finally we fit the model, and show the summary of the model fit:
simple_fit = simple_model.fit()
simple_fit.summary()
| Dep. Variable: | Creatinine | R-squared: | 0.716 |
|---|---|---|---|
| Model: | OLS | Adj. R-squared: | 0.709 |
| Method: | Least Squares | F-statistic: | 103.1 |
| Date: | Thu, 17 Oct 2024 | Prob (F-statistic): | 9.33e-13 |
| Time: | 22:46:57 | Log-Likelihood: | -95.343 |
| No. Observations: | 43 | AIC: | 194.7 |
| Df Residuals: | 41 | BIC: | 198.2 |
| Df Model: | 1 | ||
| Covariance Type: | nonrobust |
| coef | std err | t | P>|t| | [0.025 | 0.975] | |
|---|---|---|---|---|---|---|
| Intercept | -0.0849 | 0.668 | -0.127 | 0.899 | -1.434 | 1.264 |
| Urea | 0.0552 | 0.005 | 10.155 | 0.000 | 0.044 | 0.066 |
| Omnibus: | 2.409 | Durbin-Watson: | 1.303 |
|---|---|---|---|
| Prob(Omnibus): | 0.300 | Jarque-Bera (JB): | 1.814 |
| Skew: | 0.503 | Prob(JB): | 0.404 |
| Kurtosis: | 3.043 | Cond. No. | 236. |
Notes:
[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.
Notice that the coeff column towards the bottom of this output. Sure enough,
StatsModels is doing the same calculation as linregress, and getting the same
answer as minimize with our least-squares criterion. The ‘Intercept’ and
slope for ‘Urea’ are the same as those we have already seen with the other
methods.
Statsmodels where columns have spaces#
As a side-note, you have to do some extra work to tell Statsmodels formulae about column names with spaces and other characters that would make the column names invalid as variable names.
For example, let’s say we were using the original DataFrame ckd. We want to use Statsmodels to find the best line to predict 'Serum Creatinine' values from the 'Blood Urea' values. These were the original column names. We could try this:
# This generates an error, because the Statsmodels formula interface
# needs column names that work as variable names.
another_model = smf.ols(formula="Serum Creatinine ~ Blood Urea",
data=ckd)
Traceback (most recent call last):
File /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/IPython/core/interactiveshell.py:3577 in run_code
exec(code_obj, self.user_global_ns, self.user_ns)
Cell In[16], line 3
another_model = smf.ols(formula="Serum Creatinine ~ Blood Urea",
File /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/statsmodels/base/model.py:203 in from_formula
tmp = handle_formula_data(data, None, formula, depth=eval_env,
File /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/statsmodels/formula/formulatools.py:63 in handle_formula_data
result = dmatrices(formula, Y, depth, return_type='dataframe',
File /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/patsy/highlevel.py:309 in dmatrices
(lhs, rhs) = _do_highlevel_design(formula_like, data, eval_env,
File /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/patsy/highlevel.py:164 in _do_highlevel_design
design_infos = _try_incr_builders(formula_like, data_iter_maker, eval_env,
File /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/patsy/highlevel.py:66 in _try_incr_builders
return design_matrix_builders([formula_like.lhs_termlist,
File /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/patsy/build.py:689 in design_matrix_builders
factor_states = _factors_memorize(all_factors, data_iter_maker, eval_env)
File /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/patsy/build.py:354 in _factors_memorize
which_pass = factor.memorize_passes_needed(state, eval_env)
File /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/patsy/eval.py:478 in memorize_passes_needed
subset_names = [name for name in ast_names(self.code)
File /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/patsy/eval.py:478 in <listcomp>
subset_names = [name for name in ast_names(self.code)
File /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/site-packages/patsy/eval.py:109 in ast_names
for node in ast.walk(ast.parse(code)):
File /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/ast.py:50 in parse
return compile(source, filename, mode, flags,
File <unknown>:1
Serum Creatinine
^
SyntaxError: invalid syntax
The solution is to use the Q()
(Quote)
function in your formula. It tells Statsmodels that you mean the words ‘Serum’
and ‘Creatinine’ to be one thing: ‘Serum Creatinine’ - the name of the column.
another_model = smf.ols(formula="Q('Serum Creatinine') ~ Q('Blood Urea')", data=ckd)
another_fit = another_model.fit()
another_fit.summary()
| Dep. Variable: | Q('Serum Creatinine') | R-squared: | 0.803 |
|---|---|---|---|
| Model: | OLS | Adj. R-squared: | 0.802 |
| Method: | Least Squares | F-statistic: | 635.8 |
| Date: | Thu, 17 Oct 2024 | Prob (F-statistic): | 6.65e-57 |
| Time: | 22:46:58 | Log-Likelihood: | -272.97 |
| No. Observations: | 158 | AIC: | 549.9 |
| Df Residuals: | 156 | BIC: | 556.1 |
| Df Model: | 1 | ||
| Covariance Type: | nonrobust |
| coef | std err | t | P>|t| | [0.025 | 0.975] | |
|---|---|---|---|---|---|---|
| Intercept | -0.8707 | 0.163 | -5.338 | 0.000 | -1.193 | -0.548 |
| Q('Blood Urea') | 0.0582 | 0.002 | 25.215 | 0.000 | 0.054 | 0.063 |
| Omnibus: | 46.000 | Durbin-Watson: | 1.460 |
|---|---|---|---|
| Prob(Omnibus): | 0.000 | Jarque-Bera (JB): | 148.152 |
| Skew: | 1.095 | Prob(JB): | 6.75e-33 |
| Kurtosis: | 7.208 | Cond. No. | 106. |
Notes:
[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.
Multiple regression, in steps#
Now we move on to trying to predict the Creatinine using the Urea and the
Hemoglobin. The Urea values and Hemoglobin values contain different
information, so both values may be useful in predicting the Creatinine.
One way to use both values is to use them step by step - first use Urea, and
then use Hemoglobin.
First we predict the Creatinine using just the straight-line relationship we
have found for Urea.
# Use the RMSE line; but all our methods gave the same line.
intercept, slope = min_res.x
creat_predicted = intercept + slope * ckdp['Urea']
errors = ckdp['Creatinine'] - creat_predicted
# Show the first five errors
errors.head()
0 0.791367
1 1.374032
2 -0.529601
3 -0.786857
4 -4.190885
dtype: float64
The errors are the distances between the values predicted by the line, and the actual values.
scatter_errors(ckdp['Urea'], ckdp['Creatinine'], intercept, slope)
np.float64(2.2218544954119746)
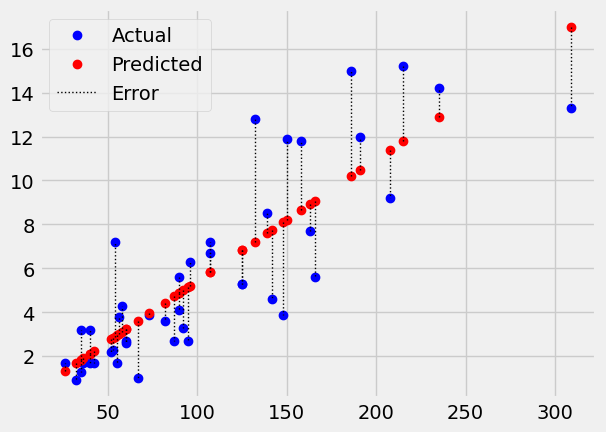
We can also call these errors residuals in the sense they are the error that
remains after removing the (straight-line) effect of Urea.
# We can also call the errors - residuals.
residuals = errors
The remaining root mean square error is:
# Root mean square error
np.sqrt(np.mean(residuals ** 2))
np.float64(2.2218544954119746)
Now we want to see if we can predict these residuals with the Hemoglobin
values. Let’s use these residuals as our new y values, and fit a predicting
line using Hemoglobin.
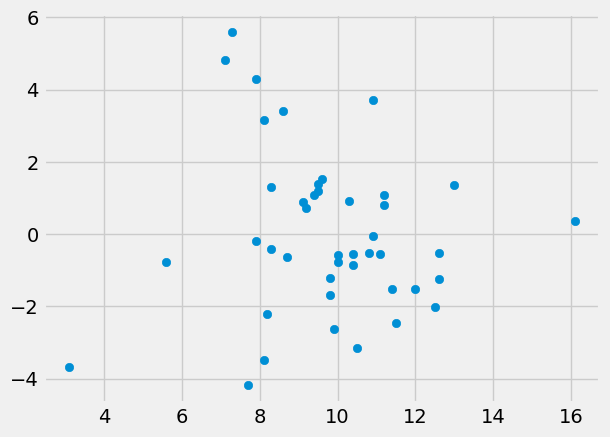
First plot the residuals (y) against the Hemoglobin (x):
plt.scatter(ckdp['Hemoglobin'], residuals)
<matplotlib.collections.PathCollection at 0x7f0263f18d50>
Then fit a line:
min_rmse_hgb = minimize(rmse_any_line,
initial_guess,
args=(ckdp['Hemoglobin'], residuals),
method='powell')
min_rmse_hgb
message: Optimization terminated successfully.
success: True
status: 0
fun: 2.221655830951226
x: [-2.462e-06 -2.960e-03]
nit: 1
direc: [[ 1.000e+00 0.000e+00]
[ 0.000e+00 1.000e+00]]
nfev: 21
The results from minimize show that the line relating Hemoglobin and the
residuals has a negative slope, as we would expect; more severe kidney disease
leads to lower hemoglobin and higher creatinine. The root mean square error
has hardly changed, suggesting that Hemoglobin does not predict much, once we
have allowed for the predictions using Urea.
Multiple regression in one go#
The other way to approach this problem is multiple regression. In multiple
regression, we use multiple columns of data at the same time to predict our
measure of interest — in this case — the Creatinine values.
In simple regression, we are using a single column of predicting values — in
our case, the Urea values — to predict the measure of interest
(Creatinine). We had to find the best pair of parameters — the intercept
(call this c) and the slope for the single column of predicting values (call
this s).
c, s = min_res.x
print('Intercept is', c)
print('Slope is', s)
Intercept is -0.08491110302073984
Slope is 0.05524186243162104
In multiple regression, we have more than one column of predicting values. For
each, we calculate a matching slope. In our new case here, the two columns of
predicting values are Urea and Hemoglobin. We therefore have to find:
An intercept. Call this
c_mto distinguish it from the intercept we found in simple regression.A slope for the line relating
UreatoCreatinine. Call thiss1.A slope for the line relating
HemoglobintoCreatinine. Call thiss2.
In the simple regression case, we had to search many intercepts and many slopes
to find the intercept, slope (c, s) pair, that gives the lowest cost function
value.
In our new case of multiple regression, we have to search many intercept, Urea
slope and Creatinine slope triplets (c_m, s1, s2) to minimize the cost
function.
For the simple case, when predicting Creatinine from Urea, we got the
predicted values by starting with the intercept c, then adding the result of
multiplying the slope for Urea (s) by the Urea values.
simple_predictions = c + s * ckdp['Urea']
The slope for Urea s gives the scaled amount of Urea to add to the
prediction.
When we have two predictors, Urea and Hemoglobin, we start with the intercept c_m, then add the result of multiplying the slope
for Urea by the Urea values, and add the result of multiplying the slope
for Hemoglobin by the Hemoglobin values. The calculation is:
The new multiple regression intercept (
c_m) plusThe
Ureaslopes1timesUreaplusThe Hemoglobin slope times Hemoglobin:
Let’s make an initial guess at the three parameters:
guessed_c_m = 0
guessed_s1 = 0.05
guessed_s2 = 0.1
The predictions are therefore:
predictions = (guessed_c_m +
guessed_s1 * ckdp['Urea'] +
guessed_s2 * ckdp['Hemoglobin'])
The root mean square error for these three parameters are:
errors = ckdp['Creatinine'] - predictions
# RMSE result
np.sqrt(np.mean(errors ** 2))
np.float64(2.3445002442629086)
Here is a function to calculate the root mean squared error for these three parameters:
def rmse_three_params(c_s_s, x1_values, x2_values, y_values):
c, s1, s2 = c_s_s
predictions = c + s1 * x1_values + s2 * x2_values
errors = y_values - predictions
return np.sqrt(np.mean(errors ** 2))
We repeat the RMSE calculation we did above, using the new function:
rmse_three_params([guessed_c_m, guessed_s1, guessed_s2],
ckdp['Urea'], ckdp['Hemoglobin'], ckdp['Creatinine'])
np.float64(2.3445002442629086)
Here we calculate the root mean square error for an intercept of 1, and slopes
for Urea and Hemoglobin of 0 and 0.
rmse_three_params([1, 0, 0],
ckdp['Urea'], ckdp['Hemoglobin'], ckdp['Creatinine'])
np.float64(6.28908245609285)
Now we can get minimize to find the intercept and two slopes that minimize the
root mean square error (and the sum of squared error):
min_css = minimize(rmse_three_params, [0, 0, 0], method='powell',
args=(ckdp['Urea'], # This will become x1_values above.
ckdp['Hemoglobin'], # This will become x2_values above.
ckdp['Creatinine'] # This will become y_values above.
))
min_css
message: Optimization terminated successfully.
success: True
status: 0
fun: 2.2155376442850536
x: [ 1.025e+00 5.345e-02 -9.457e-02]
nit: 6
direc: [[ 1.000e+00 0.000e+00 0.000e+00]
[ 4.412e-01 1.711e-02 -2.049e-01]
[-5.009e+00 1.058e-02 3.779e-01]]
nfev: 193
Just as for the simple regression case, and linregress, we can get our
parameters by calculation directly, for this case where we are using
least-squares as our criterion.
Don’t worry about the details of the function below. It contains the matrix
calculation to give us the same answer as minimize above, as long as we are
minimizing the root mean square error (or sum of squared error) for an
intercept and two slopes.
def multiple_regression_triple(x1_values, x2_values, y_values):
""" linregress equivalent for intercept and two slopes
Parameters
----------
x1_values: array-like, shape (n,)
First sequence (such as an array) of values to predict `y_values`.
x2_values: array-like, shape (n,)
First sequence (such as an array) of values to predict `y_values`.
y_values : array-like, shape (n,)
Values to be predicted.
Returns
-------
params : array, shape (3,)
Least-squares fit parameters, where first parameter is intercept value,
second is slope for `x1_values`, and third is slope for `x2_values`.
"""
intercept_col = np.ones(len(y_values))
X = np.column_stack([intercept_col, x1_values, x2_values])
return np.linalg.pinv(X) @ y_values
This function gives the same result as we got from minimize.
params = multiple_regression_triple(
ckdp['Urea'], ckdp['Hemoglobin'], # x values.
ckdp['Creatinine']) # y values.
params
array([ 1.02388056, 0.05345685, -0.09432084])
Finally in this section, let’s see StatsModels in action, to do the same calculation.
Here we specify that we want to fit a linear model to Creatinine as a
function of Urea and as a function of Hemoglobin. This has the same
meaning as above; that we will simultaneously fit the intercept, the Urea
slope and the Hemoglobin slope.
multi_model = smf.ols(formula="Creatinine ~ Urea + Hemoglobin", data=ckdp)
multi_fit = multi_model.fit()
multi_fit.summary()
| Dep. Variable: | Creatinine | R-squared: | 0.717 |
|---|---|---|---|
| Model: | OLS | Adj. R-squared: | 0.703 |
| Method: | Least Squares | F-statistic: | 50.70 |
| Date: | Thu, 17 Oct 2024 | Prob (F-statistic): | 1.08e-11 |
| Time: | 22:46:58 | Log-Likelihood: | -95.221 |
| No. Observations: | 43 | AIC: | 196.4 |
| Df Residuals: | 40 | BIC: | 201.7 |
| Df Model: | 2 | ||
| Covariance Type: | nonrobust |
| coef | std err | t | P>|t| | [0.025 | 0.975] | |
|---|---|---|---|---|---|---|
| Intercept | 1.0239 | 2.416 | 0.424 | 0.674 | -3.859 | 5.907 |
| Urea | 0.0535 | 0.007 | 8.049 | 0.000 | 0.040 | 0.067 |
| Hemoglobin | -0.0943 | 0.197 | -0.478 | 0.635 | -0.493 | 0.305 |
| Omnibus: | 1.746 | Durbin-Watson: | 1.246 |
|---|---|---|---|
| Prob(Omnibus): | 0.418 | Jarque-Bera (JB): | 1.326 |
| Skew: | 0.430 | Prob(JB): | 0.515 |
| Kurtosis: | 2.960 | Cond. No. | 851. |
Notes:
[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.
Notice again that StatsModels is doing the same calculation as above, and
finding the same result as does minimize.
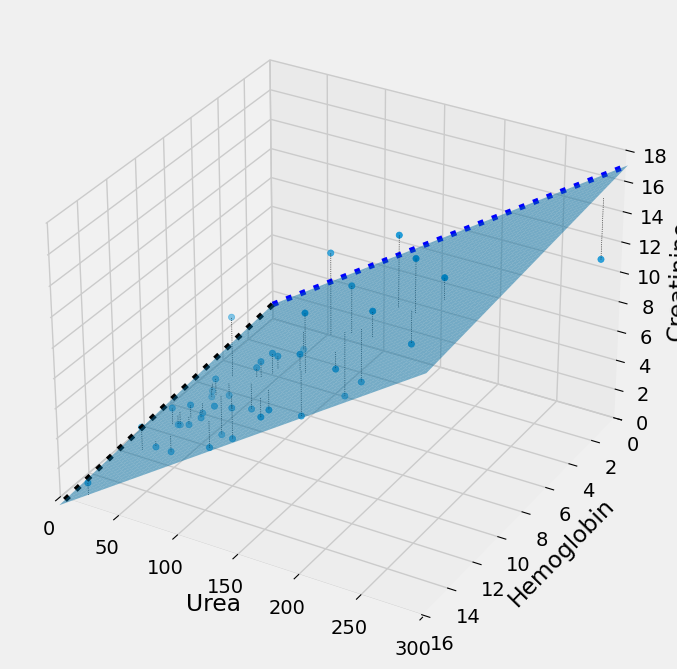
Multiple regression in 3D#
It can be useful to use a 3D plot to show what is going on here. minimize
and the other methods are finding these three parameters simultaneously:
An intercept;
A slope for
UreaA slope for
Hemoglobin.
The plot below shows what this looks like, in 3D. Instead of the 2D case,
where we are fitting the y data values (Creatinine) with a single straight
line, here we are fitting the y data values with two straight lines. In 3D
these two straight lines form a plane, and we want the plane such that the sum
of squares of the distance of the y values from the plane (plotted) is as small
as possible. minimize will change the intercept and the two slopes to move
this plane around until it has minimized the error.
Show code cell content
# Run this cell.
import mpl_toolkits.mplot3d # (for Matplotlib < 3.2)
ax = plt.figure(figsize=(8,8)).add_subplot(111, projection='3d')
ax.scatter(ckdp['Urea'],
ckdp['Hemoglobin'],
ckdp['Creatinine']
)
ax.set_xlabel('Urea')
ax.set_ylabel('Hemoglobin')
ax.set_zlabel('Creatinine')
intercept, urea_slope, hgb_slope = min_css.x
mx_urea, mx_hgb, mx_creat = 300, 16, 18
ax.plot([0, mx_urea],
[intercept, intercept + urea_slope * mx_urea],
0,
zdir='y', color='blue', linestyle=':')
mx_hgb = ckdp['Hemoglobin'].max()
ax.plot([0, mx_hgb],
[intercept, intercept + hgb_slope * mx_hgb],
0,
zdir='x', color='black', linestyle=':')
# Plot the fitting plane.
plane_x = np.linspace(0, mx_urea, 50)
plane_y = np.linspace(0, mx_hgb, 50)
X, Y = np.meshgrid(plane_x, plane_y)
Z = intercept + urea_slope * X + hgb_slope * Y
ax.plot_surface(X, Y, Z, alpha=0.5)
# Plot lines between each point and fitting plane
for i, row in ckdp.iterrows():
x, y, actual = row['Urea'], row['Hemoglobin'], row['Creatinine']
fitted = intercept + x * urea_slope + y * hgb_slope
ax.plot([x, x], [y, y], [fitted, actual],
linestyle=':',
linewidth=0.5,
color='black')
# Set the axis limits (and reverse y axis)
ax.set_xlim(0, mx_urea)
ax.set_ylim(mx_hgb, 0)
ax.set_zlim(0, mx_creat);
And even more parameters#
At the top of this page, we started by finding two parameters:
intercept
slope for
Urea
Then we extended this to three parameters (two slopes):
intercept
slope for
Ureaslope for
Hemoglobin
To get the predicted values for the three-parameter model we take
the intercept plus
the slope for
Ureatimes theUreavalues plusthe slope for
Hemoglobintimes theHemoglobinvalues.
In fact we can extend this idea further by adding more columns of values, and more slopes. For example, imagine I want to be able to send a whole DataFrame of columns to the cost function, each with its matching slope, I could do this:
def rmse_n_params(params, attributes, y_values):
""" RMSE for intercept, slopes model of `y_values` using `attributes`
Parameters
----------
params : array-like, shape (p + 1,)
Intercept (``params[0]``) and slopes ``params[1:]``, with one slope for each column in `attributes`.
attributes : pd.Dataframe, shape (n, p)
2D DataFrame, with one column per predicting parameter.
y_values : array-like, shape (n,)
1-dimensional array containing values to be predicted.
Returns
-------
rmse : float
Root mean squared error when predicting `y_values` from `c + params[1] *
attributes.iloc[:, 0] + params[2] * attributes.iloc[:,2] ...`
"""
c = params[0] # The intercept
slopes = params[1:] # One slope for each column in attributes.
predictions = c # Start with intercept.
for col_no in np.arange(len(slopes)):
col = attributes.iloc[:, col_no] # Get predictor.
col_contribution = slopes[col_no] * col # Scale predictor.
predictions = predictions + col_contribution # Add scaled predictor.
errors = y_values - predictions
return np.sqrt(np.mean(errors ** 2))
First we show off the more general function by re-doing our two-parameter calculation:
# Two columns of attributes
attributes2 = ckdp.loc[:, ['Urea', 'Hemoglobin']]
# Recalculate the RMSE
rmse_n_params([1, 0, 0], attributes2, ckdp['Creatinine'])
np.float64(6.28908245609285)
Using the more general function for the two-parameter optimization:
min_css = minimize(rmse_n_params, [1, 0, 0], method='powell',
args=(attributes2, ckdp['Creatinine']))
min_css
message: Optimization terminated successfully.
success: True
status: 0
fun: 2.2155377197879225
x: [ 1.025e+00 5.345e-02 -9.459e-02]
nit: 6
direc: [[ 1.000e+00 0.000e+00 0.000e+00]
[ 4.416e-01 1.713e-02 -2.050e-01]
[-5.007e+00 1.057e-02 3.774e-01]]
nfev: 194
We can add as many columns as we want, and ask minimize to find the best slopes for each column. Here is the result with three columns:
attributes3 = ckdp.loc[:, ['Urea', 'Hemoglobin', 'WBC']]
min_css3 = minimize(rmse_n_params, [1, 0, 0, 0], method='powell',
args=(attributes3, ckdp['Creatinine']))
min_css3
message: Optimization terminated successfully.
success: True
status: 0
fun: 2.199779156129968
x: [ 1.807e+00 5.222e-02 -9.702e-02 -5.987e-05]
nit: 7
direc: [[ 1.000e+00 0.000e+00 0.000e+00 0.000e+00]
[ 5.427e-02 7.640e-05 6.069e-02 -5.264e-05]
[ 6.375e-01 1.695e-02 -2.034e-01 -2.152e-05]
[-4.795e+00 9.883e-03 3.195e-01 5.090e-05]]
nfev: 331
We can also generalize the mathematical calculation to solve the same problem, as long as we do want the best parameters for least-squares problems:
def multiple_regression_matrix(attributes, y_values):
""" linregress equivalent for multiple slopes
Parameters
----------
attributes : array-like, shape (n, p)
2-dimensional array-like (such as a DataFrame) where each column is a
regressor (covariate), to predict corresponding `y_values`.
y_values : array-like, shape (n,)
Values to be predicted.
Returns
-------
params : array, shape (p + 1,)
Least-squares fit parameters, where first parameter is intercept value,
second is slope for first column in `attributes`, third is slope for
second column in `attributes`, and so on.
"""
intercept_col = np.ones(len(y_values))
X = np.column_stack([intercept_col, attributes])
return np.linalg.pinv(X) @ y_values
We get the same result from this calculation as we did from minimize.
multiple_regression_matrix(attributes3, ckdp['Creatinine'])
array([ 1.80554571, 0.05222674, -0.09691451, -0.00005944])